Difference Between Sanger Sequencing and Pyrosequencing
Key Difference – Sanger Sequencing vs Pyrosequencing
DNA sequencing is very important for DNA analysis since knowledge of the correct nucleotides arrangement on a particular DNA region reveals many important information about it. There are different DNA sequencing methods. Sanger sequencing and Pyrosequencing are two different DNA sequencing methods widely used in Molecular Biology. The key difference between Sanger sequencing and Pyrosequencing is that Sanger sequencing uses dideoxynucleotides to terminate the synthesis of DNA to read the nucleotide sequence while pyrosequencing detects the pyrophosphate release by incorporating the nucleotides and synthesizing the complementary sequence to read the precise order of the sequence.
CONTENTS
1. Overview and Key Difference
2. What is Sanger Sequencing
3. What is Pyrosequencing
4. Side by Side Comparison – Sanger Sequencing vs Pyrosequencing
5. Summary
What is Sanger Sequencing?
Sanger sequencing is a first generation DNA sequencing method developed by Frederick Sanger and his colleges in 1977. It is also known as Chain Termination Sequencing or Dideoxy sequencing since it is based on chain termination by dideoxynucleotides (ddNTPs). This method was widely used for more than 30 years until the New Generation Sequencing (NGS) was developed. Sanger sequencing technique enabled the discovery of the correct nucleotide order or the attachment of a particular DNA fragment. It is based on the selective incorporation of ddNTPs and termination of DNA synthesis during the in vitro DNA replication. The absence of 3’ OH groups to continue phosphodiester bond formation between adjacent nucleotides is a unique feature of ddNTPs. Hence, once the ddNTP is attached, chain elongation ceases and terminates from that point. There are four ddNTPs – ddATP, ddCTP, ddGTP and ddTTP – used in Sanger sequencing. These nucleotides stop the DNA replication process when they are incorporated into the growing strand of DNA and result in varying lengths of short DNA. Capillary gel electrophoresis is used to organize these short DNA strands by their sizes on a gel as shown in Figure 01.
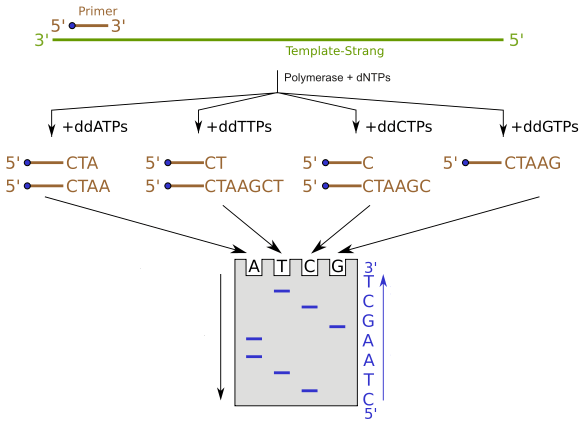
Figure 1: Capillary gel electrophoresis of synthesized short DNA
For in vitro replication of DNA, few requirements should be provided. They are DNA polymerase enzyme, template DNA, oligonucleotide primers, and deoxynucleotides (dNTPs). In Sanger sequencing, DNA replication is performed in four separate test tubes along with four types of ddNTPs separately. Deoxynucleotides are not totally replaced by the respective ddNTPs. A mixture of the particular dNTP (for example; dATP + ddATP) is included in the tube and replicated. Four separate tube products are run on a gel in four separate wells. Then by reading the gel, the sequence can be constructed as shown in Figure 02.
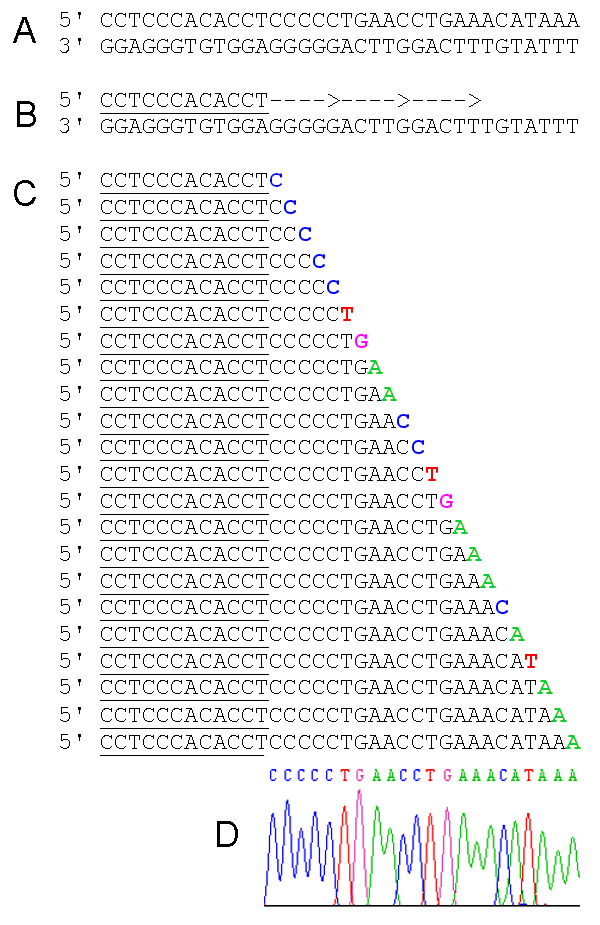
Figure 02: Sanger sequencing
Sanger sequencing is an important technique that helps in many areas of molecular biology. Human genome project was successfully completed with the aid of Sanger sequencing-based methods. Sanger sequencing is also useful in target DNA sequencing, cancer and genetic disease research, gene expression analysis, human identification, pathogen detection, microbial sequencing etc.
There are several disadvantages of Sanger sequencing:
- The length of the DNA being sequenced cannot be longer than 1000 base pairs
- Only one strand can be sequenced at a time.
- The process is time-consuming and expensive.
Therefore, new advanced sequencing techniques were developed with time to overcome these problems. However, Sanger sequencing is still in use due to its highly accurate results up to approximately 850 base pair length fragments.
What is Pyrosequencing?
Pyrosequencing is a novel DNA sequencing technique based on the “sequencing by synthesis”. This technique relies on the detection of pyrophosphate release upon the nucleotide incorporation. The process is employed by four different enzymes: DNA polymerse, ATP sulfurylase, luciferase and apyrase and two substrates adenosine 5’ phosphosulfate (APS) and luciferin.
The process starts with the primer binding with the single-stranded DNA template and DNA polymerase starts the incorporation of nucleotides complementary to it. When the nucleotides join together (nucleic acid polymerization), it releases pyrophosphate (two phosphate groups bound together) groups and energy. Each nucleotide addition releases equimolar quantity of pyrophosphate. Pyrophosphate converts into ATP by ATP sulfurylase in the presence of substrate APS. The generated ATP drives the luciferase-mediated conversion of luciferin to oxyluciferin, producing visible light in amounts that are proportional to the amount of ATPs. Light is detected by a photon detection device or by photomultiplier and creates a pyrogram. Apyrase degrades ATP and non-incorporated dNTPs in the reaction mixture. dNTP addition is done once at a time. Since the addition of nucleotide is known according to the incorporation and detection of light, the sequence of the template can be determined. Pyrogram is used for generating the nucleotide sequence of the sample DNA as shown in Figure 03.
Pyrosequencing is very important in single nucleotide polymorphism analysis and sequencing of short stretches of DNA. The high accuracy, flexibility, ease of automation and parallel processing are the advantages of pyrosequencing over Sanger sequencing techniques.
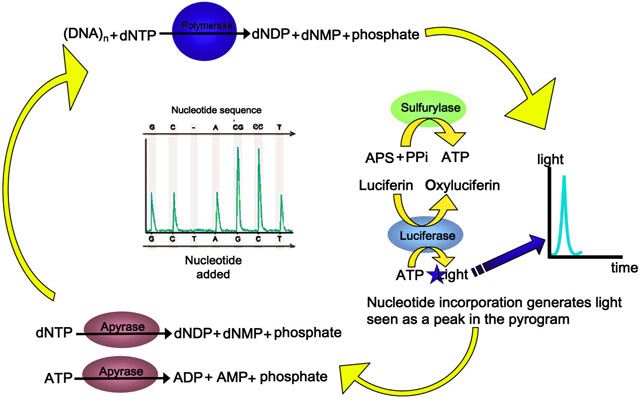
Figure 03: Pyrosequencing
What is the difference between Sanger Sequencing and Pyrosequencing?
Sanger Sequencing vs Pyrosequencing | |
| Sanger sequencing is a DNA sequencing method based on the selective incorporation of ddNTPs by DNA polymerase and chain termination. | Pyrosequencing is a DNA sequencing method based on the detection of pyrophosphate release upon the incorporation of nucleotide. |
| Use of ddNTP | |
| ddNTPs are used to terminate the DNA replication | ddNTPs are not used. |
| Enzymes involved | |
| DNA polymerase are used. | Four enzymes are used: DNA polymerase, ATP sulfurylase, Luciferase and Apyrase. |
| Substrates Used | |
| APS and Luciferin are not used. | Adenosine 5’ phosphosulfate (APS) and luciferin are used. |
| Maximum Temperature | |
| This is a slow process. | This is a fast process. |
Summary – Sanger Sequencing vs Pyrosequencing
Sanger sequencing and Pyrosequencing are two DNA sequencing methods used in molecular biology. Sanger sequencing constructs the order of the nucleotides in sequence by terminating the chain elongation while the pyrosequencing constructs the precise order of the nucleotides in sequence by incorporation of nucleotides and detecting the release of pyrophosphates. Therefore, the main difference between Sanger sequencing and Pyrosequencing is that Sanger sequencing works on sequencing by chain termination while pyrosequencing works on sequencing by synthesis.
Reference:
1. Fakruddin, Md, and Abhijit Chowdhury. “Pyrosequencing-An Alternative to Traditional Sanger Sequencing.” American Journal of Biochemistry and Biotechnology. Science Publications, 02 Mar. 2012. Web. 28 Feb. 2017.
2. “Sanger sequencing.” Sanger sequencing – ScienceDirect Topics. N.p., n.d. Web. 28 Feb. 2017
Image Courtesy:
1. “Didesoxy-Methode”By Christoph Goemans (modifiziert) – Dr. Norman Mauder, auf Basis einer Datei von Christoph Goemans (CC BY-SA 3.0) via Commons Wikimedia
2. “Sanger-DNA-seq” By Enzo at the Polish language Wikipedia (CC BY-SA 3.0) via Commons Wikimedia
3. “Pyrosequencing” by microbiologybytes (CC BY-SA 2.0) via Flickr
ncG1vNJzZmivp6x7pbXFn5yrnZ6YsqOx07CcnqZemLyue8OinZ%2Bdopq7pLGMm5ytr5Wau26%2FwKeenqpdqLKywcSnmqKml2Kur7CMr6pmqKmnvLSx0K6cp5uZo7Rw